Creating 96-Well Plates in R
This week I did some virus neutralization test and I was wondering if there is any way to show the results in 96-well plate format. I was trying to find if there is any R package that can do this. Actually there is one (or might be more). I found a package called platetools that can plot values in plate format.
Let’s see how I used this package.
R Markdown
First load all the required packages
library(ggplot2)
library(platetools)
library(viridis)
## Loading required package: viridisLitePrepare your datasets.I prepared in R as shown below.
data=read.csv("df.csv")
data
## X vals well
## 1 1 1 A01
## 2 2 0 A02
## 3 3 0 A03
## 4 4 1 A04
## 5 5 0 A05
## 6 6 0 A06
## 7 7 0 A07
## 8 8 0 A08
## 9 9 0 A09
## 10 10 0 A10
## 11 11 0 A11
## 12 12 1 A12
## 13 13 1 B01
## 14 14 0 B02
## 15 15 0 B03
## 16 16 1 B04
## 17 17 0 B05
## 18 18 0 B06
## 19 19 0 B07
## 20 20 0 B08
## 21 21 0 B09
## 22 22 0 B10
## 23 23 0 B11
## 24 24 1 B12
## 25 25 1 C01
## 26 26 0 C02
## 27 27 0 C03
## 28 28 1 C04
## 29 29 0 C05
## 30 30 0 C06
## 31 31 0 C07
## 32 32 0 C08
## 33 33 0 C09
## 34 34 0 C10
## 35 35 1 C11
## 36 36 0 C12
## 37 37 1 D01
## 38 38 0 D02
## 39 39 0 D03
## 40 40 0 D04
## 41 41 0 D05
## 42 42 0 D06
## 43 43 0 D07
## 44 44 0 D08
## 45 45 0 D09
## 46 46 0 D10
## 47 47 1 D11
## 48 48 0 D12
## 49 49 0 E01
## 50 50 0 E02
## 51 51 0 E03
## 52 52 0 E04
## 53 53 0 E05
## 54 54 0 E06
## 55 55 0 E07
## 56 56 0 E08
## 57 57 0 E09
## 58 58 0 E10
## 59 59 1 E11
## 60 60 0 E12
## 61 61 0 F01
## 62 62 0 F02
## 63 63 0 F03
## 64 64 0 F04
## 65 65 0 F05
## 66 66 0 F06
## 67 67 0 F07
## 68 68 0 F08
## 69 69 0 F09
## 70 70 0 F10
## 71 71 1 F11
## 72 72 0 F12
## 73 73 0 G01
## 74 74 0 G02
## 75 75 0 G03
## 76 76 0 G04
## 77 77 0 G05
## 78 78 0 G06
## 79 79 0 G07
## 80 80 0 G08
## 81 81 0 G09
## 82 82 0 G10
## 83 83 1 G11
## 84 84 0 G12
## 85 85 0 H01
## 86 86 0 H02
## 87 87 0 H03
## 88 88 0 H04
## 89 89 0 H05
## 90 90 0 H06
## 91 91 0 H07
## 92 92 0 H08
## 93 93 0 H09
## 94 94 0 H10
## 95 95 1 H11
## 96 96 0 H12Finally plot your data
plate_id <- rep(c("My Plate"), each = 96)
z_grid(data = data$vals,
well = data$well,
plate_id = plate_id) +
ggtitle("Virus Neutralization Test")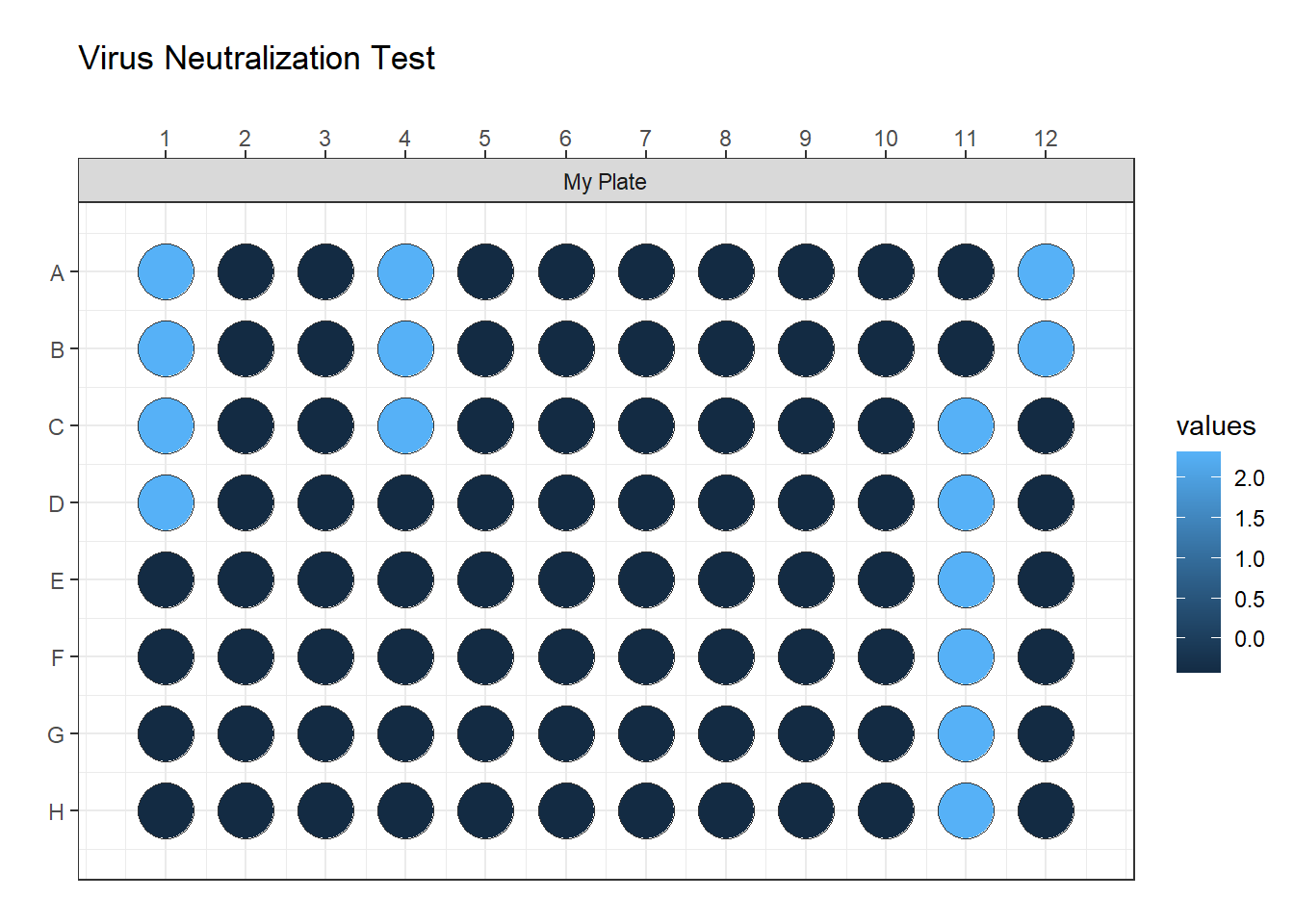
Here, the label is not useful for me because I am interested only in positive and negative wells. But it can be useful in plotting ELISA plates.